Molecular Analysis of Plant-Pathogen Interactions

One main focus of research in our lab is to understand the molecular mechanisms regulating plant-pathogen interactions. Infection of host plants by pathogens results in one of two possible outcomes: disease (compatible interaction) or resistance (incompatible interaction). Incompatible interaction is usually associated with the appearance of necrotic flecks containing dead plant cells at the sites of pathogen infection and is called the hypersensitive response (HR).
The initiation of HR (a type of programmed cell death) requires an elicitor produced by the pathogen, its recognition by the plant, signal transduction and finally activation of several plant defense genes. In many cases, HR is followed by development of long lasting systemic resistance and is know as systemic acquired resistance (SAR). SAR provides resistance to subsequent attacks by a wide variety of pathogens. While cell death associated with HR is controlled by a genetic program in the plant and requires active host participation, the mechanism of cell death during disease is not know. The long-term goal of our research is to use the tools of functional genomics to better understand the genetic and molecular mechanisms regulating activation of HR, SAR, and disease-associated host cell death during pathogen infections. We have used both forward and reverse genetic approaches to identify the plant genes involved in regulating defense against pathogens:
1. Analysis of Arabidopsis mutants with altered response to pathogen infections (forward genetics)
We have used genetic screens to identify the Arabidopsis mutants that display altered response to pathogen infections. These mutants are likely to represent genes involved in regulating plant’s response to pathogen infections. Among the mutants identified in these screens, we have analyzed two mutants in detail: a) hrl1, which mimics plant’s response to avirulent pathogens, and b) dll1, which mimics plant’s response to virulent pathogens. These mutants are briefly discussed below:
a) Characterization of hrl1 mutant

The hrl1 mutant of Arabidopsis, spontaneously develops HR-like lesions, constitutively expresses defense-related genes, and exhibits enhanced resistance to virulent strains of bacterial and fungal pathogens. Detailed genetic and molecular analysis of this mutant revealed that multiple pathogen defense pathways are constitutively activated in this mutant. Interestingly, in contrast to previous reports, we found that the signaling components of salicylic acid (SA) and ethylene/jasmonic acid (ET/JA) defense pathways function synergistically to regulate the expression of SA and ET/JA regulated defense genes and resistance against virulent pathogens in hrl1. Our studies suggest that endogenous defense signals enable plants to fine tune their transcriptional read out against a wide variety of pathogens through synergistic or antagonistic regulation of multiple defense pathways (read more). Furthermore, we found that constitutive SAR in the hrl1 or pre-induced SAR in the wild type plants suppresses HR-associated cell death induced by avirulent pathogens. Our studies suggest that once the defense is induced in response to pathogen infection, plants use a feedback regulatory system to suppress further HR-associated cell death. This may be a way for plants to prevent excessive defense-associated cell death once the SAR is induced (read more). Map-based cloning of HRL1 gene is in progress. In addition, we are using hrl1 and “defense/stress microarrays” as tools to analyze signaling components regulating cross talk between multiple pathogen defense pathways and to understand the mechanisms regulating suppression of HR-associated cell death by SAR.
b) Characterization of dll1

The dll1 (disease-like lesions) is a novel gain-of-function mutant of Arabidopsis, which spontaneously develops lesions mimicking disease, constitutively expresses all tested molecular and biochemical markers associated with disease. However, despite constitutive expression of multiple pathogen defense responses, dll1 does not suppress the growth of virulent pathogens. Expression of defense responses and yet failure to suppress the growth of virulent pathogens is a hallmark of disease. Our results suggest that the host may, at least in part, activate cell death during disease and DLL1 may positively regulate this process. Thus dll1 provides a valuable genetic tool to understand the mechanisms regulating host cell death during disease. Furthermore, our results suggest a significant overlap exists between signaling mechanisms regulating disease and resistance responses (read more). Map-based cloning of DLL1 gene is in progress. In addition, we are using dll1 and virulent pathogen-infected wild type plants together with ‘defense/stress microarrays’ to identify the plant genes that regulate host cell death during disease.
2. Functional genomics of HR, disease and SAR using “defense/stress microarrays” (reverse genetics)
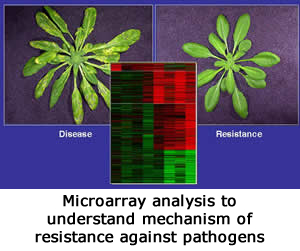 This involves identification of genes differentially expressed during disease, HR and SAR responses to pathogens their detailed characterization.
This involves identification of genes differentially expressed during disease, HR and SAR responses to pathogens their detailed characterization.
a) Identification of differentially expressed genes
To identify differentially expressed genes, we have used the Suppression Subtractive Hybridization method (SSH) to construct cDNA libraries enriched for genes that are differentially expressed in response to virulent and avirulent strains of P. syringae, Alternaria brassicicola (a fungal pathogen), P. fluorescence (root colonizing bacteria induces ISR), P. parasitica (fungal pathogen), ozone (reactive oxygen species generator), methyl jasmonate (inducer of ET/JA defense pathway), salicylic acid (activator of SAR). From these screens, we identified more than 1000 genes that are differentially expressed in response to one or more of these stresses. Among these, only 20% have been previously associated with defense against pathogens. Based on homology to genes with known functions, 72% genes were predicted to encode proteins with a variety of cellular functions such as signal transduction, transcriptional regulation, metabolic processes etc. About 17% of these genes have no match to any EST in the public database. The genes identified in these studies provide a very useful resource to understand the molecular events involved in activating defense in plants against pathogens (read more). We used these and other stress/defense related genes to construct “defense/stress cDNA microarrays”. These microarrays were used to study temporal expression profiles of the genes on the microarray in response to: a) variety of biotic stresses (virulent and avirulent strains of bacterial, viral and fungal pathogens and insects), b) variety of abiotic stresses (heat, cold, drought, salinity, wounding, osmotic stress etc.), b) plant hormones (salicylic acid, ethylene, abscisic acid, gibberellins, cytokinins, auxins, brassinosteroid), and c) paraquat, 3AT, glucose-glucose oxidase, xanthine-xanthine oxidase (inducers of oxidative stress). Results of these experiments together with tools of bioinformatics were used to construct a searchable database Arabidopsis Stress Microarray Database to search for genes that are unique to each stress or participate in regulating multiple stresses. This database will be posted on the internet after the manuscript describing these results has been published (see the attached front page of this database). This analysis identified signaling pathways that are unique to each stress and common to multiple stresses. The results from these studies are being used to understand underlying signaling events involved in regulating plant’s response to pathogens.
b) Functional analysis of genes differentially expressed during defense activation
To understand the biological function of defense-related genes identified in the studies described above, we have selected several genes that are predicted to encode novel transcription factors or signaling proteins to construct transgenic plants that over- or under-express these target genes (35S promoter for over-expression and RNAi for under-expression). These include some WRKY genes, Zn finger protein, protein phosphatase 2C, lectin-like kinase, MAP kinase, Ser/Thr kinase, receptor-like kinases etc. These transgeneic plants are being used to: a) Determine the role of the target genes in regulating defense against virulent pathogen, and b) Construct gene networks regulating plant-pathogen interactions.
Following are some examples
i) Cysteine-Rich Receptor-Like Kinase 13 (CRK13)

Protein kinases play important roles in relaying information from perception of a signal to the effector genes in all organisms. Cysteine-rich Receptor-like Kinases(CRKs) constitute a subfamily of plant Receptor-Like Kinases (RLKs) with more than 40 members that contain the novel C-X8-C-X2-C motif (DUF26) in the extracellular domains. Here we report molecular characterization of one member of this gene family, CRK13. Expression of this gene is induced quicker and stronger in response to the avirulent compared to virulent strains of Pseudomonas syringae and peaks within 4 h after pathogen infection. In response to dexamethasone (DEX) treatment, plants expressing the CRK13 gene from a DEX-inducible promoter exhibited all tested features of pathogen defense activation, including rapid tissue collapse, accumulation of high levels of several defense-related gene transcripts including PR1, PR5 and ICS1, and accumulation of salicylic acid (SA). In addition, these plants suppressed growth of virulent pathogens about 20-fold compared to the wild-type Col-0. CRK13-conferred pathogen resistance is SA dependent. Gene expression analysis using custom cDNA microarrays revealed a remarkable overlap between the expression profiles of the plants overexpressing CRK13 and the plants treated with Pst DC3000 (avrRpm1). Our studies suggest that upregulation of CRK13 leads to hypersensitive response-associated cell death and induces defense against pathogens by causing increased accumulation of SA (read more).
ii) GH3-Like Defense Gene 1 (GDG1)

In Arabidopsis, GH3-like gene family consists of 19 members, several of which have been shown to adenylate the plant hormones jasmonic acid (JA), indole acetic acid (IAA) and salicylic acid (SA). In some cases this adenylation has been shown to catalyze hormone conjugation to amino acids. Here we report molecular characterization of the GH3-LIKE DEFENSE GENE 1 (GDG1), a member of the GH3-like gene family and show that GDG1 is an important component of SA-mediated defense against the bacterial pathogen Pseudomonas syringae. Expression of GDG1 is induced earlier and to a higher level in response to avirulent pathogens compared to virulent pathogens. gdg1 null mutants are compromised in several pathogen defense responses, including activation of defense genes and resistance against virulent and avirulent bacterial pathogens. Accumulation of free and glucoside-conjugated SA (SAG) in response to pathogen infection is compromised in gdg1 mutants. All defense-related phenotypes of gdg1 can be rescued by external application of SA, suggesting that gdg1 mutants are defective in the SA-mediated defense pathway(s) and that GDG1 functions upstream of SA. Our results suggest that GDG1 contributes to both basal and resistance gene-mediated inducible defenses against P. syringae (and possibly other pathogens) by playing a critical role in regulating levels of pathogen-inducible SA. GDG1 is allelic to PBS3 (avrPphB susceptible) gene (read more). Several other genes are currently being studied.
Functional Genomics of Receptor-Like Kinases in Arabidopsis
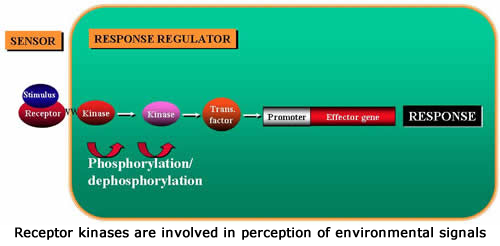
Receptor-like kinases (RLKs) are involved in signal perception through the cell surface. In Arabidopsis RLKs constitute a large family consisting of more than 600 family members. However, the function of very few of these genes is known. We have initiated a major project directed towards functional analysis of RLKs in Arabidopsis. The goal of this project is to develop Arabidopsis plants expressing chimeric RLKs, which induce visible responses (express GFP reporter gene) when they sense various chemical and biological agents. The chimeric RLKs will have the extracellular domain of the target RLK fused to the kinase domain of an RLK involved in eliciting hypersensitive response and downstream defense responses. These chimeric RLKs will be introduced in the Col-0 plants expressing GFP from the promoter of the PR-1 gene (a tightly regulated Arabidopsis defense gene). Several hundred RLK genes have been targeted for this project. Transgenic plants expressing these chimeric RLKs will constitute a “Chimeric Receptor Kit” and will be screened to identify the extracellular domain(s) involved in perception of many stimuli of interest. Recognition of the ligand by the extracellular domain of the chimeric RLK should activate the kinase, which in turn would induce HR and expression of GFP. If an RLK functions in a developmental process, transgenic plants would be expected to express GFP at that stage of development/tissue. So far we have constructed and tested more than 50 chimeric RLKs and found two that activate expression of defense genes in a predictable manner. Thus these transgenic plants should be useful to identify candidate Arabidopsis RLKs for a wide variety of biotic and abiotic environmental stimuli and possibly those involved in developmental processes.
